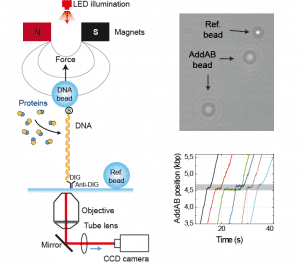
The activity of proteins that modify the extension of the DNA as a consequence of their function can be monitored using Magnetic Tweezers.
In our group we have two magnetic tweezers capable to apply forces between 0.1-70 pN and to track tens of beads with millisecond precision.
MT-related papers of the group
Coloma J, Gonzalez-Rodriguez N, Balaguer FA, Gmurczyk K, Aicart-Ramos C, Nuero ÓM, Luque-Ortega JR, Calugaru K, Lue NF, Moreno-Herrero F, Llorca O. Nucleic Acids Res. (2023). https://doi.org/10.1093/nar/gkac1261
Molecular architecture and oligomerization of Candida glabrata Cdc13 underpin its telomeric DNA-binding and unfolding activity.
Barbara Martin-Garcia*, Alejandro Martin-Gonzalez* et al. Nucleic Acids Research 14 May; gky370, https://doi.org/10.1093/nar/gky370 (2018).
The TubR-centromere complex adopts a double-ring segrosome structure in Type III partition systems.
LINK
Madariaga-Marcos et al. Nanoscale Mar 1;10(9):4579-4590 (2018). doi: 10.1039/c7nr07344e.
Force determination in lateral magnetic tweezers combined with TIRF microscopy.
LINK
Gemma LM Fisher*, Cesar L Pastrana*, Victoria A Higman* et al. eLife Dec 15;6 (2017) pii: e28086. doi: 10.7554/eLife.28086.
The structural basis for dynamic DNA binding and bridging interactions which condense the bacterial centromere.
LINK
Gilhooly et al. Nucleic Acids Research 44(6), 2727-2741 Published Online Jan 13 (2016).
Chi hotspots trigger a conformational change in the helicase-like domain of AddAB to activate homologous recombination
LINK
Gollnick et al. Small 11(11), 1273-1284 (2015), (Accepted, Oct 7, 2014). (cover article).
Probing DNA helicase kinetics with temperature controlled magnetic tweezers
LINK
Taylor*, Pastrana* et al. Nucleic Acids Research 43(2), 719-731 (2015) (Accepted, 28-Nov, 2014, Online 8 Jan 2015).
Specific and non-specific interactions of ParB with DNA: implications for chromosome segregation
LINK
Carrasco et al. DNA Repair 20, 119-129 (2014) (cover article).
Single molecule approaches to monitor the recognition and resection of double-stranded DNA breaks during homologous recombination
LINK
Carrasco et al. Proceedings of the National Academy of Sciences USA 110 (28), E2562-2571 (2013).
On the Mechanism of Recombination Hotspot Sequence Scanning by a Bacterial Helicase-Nuclease
LINK
Herrero-Galan et al. The Journal of the American Chemical Society 135(1), 122-131 (2013).
Mechanical identities of RNA and DNA double helices unveiled at the single-molecule level
LINK
Carrasco and Moreno-Herrero Enclycopedia of Life Sciences DOI:10.1002/9780470015902.a0023173 (2011).
Magnetic Tweezers
LINK
Hormeno et al. Biophysical Journal 100, 1996-2006 (2011).
Mechanical Properties of High GoC-content DNA with A-type base-stacking
LINK
Hormeno et al. Biophysical Journal 100, 2006-2015 (2011).
Condensation prevails over B-A transition in the structure of DNA at low humidity”
LINK
