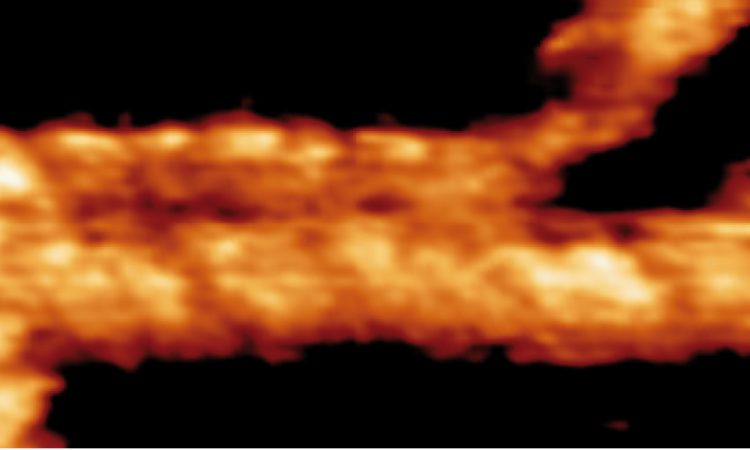
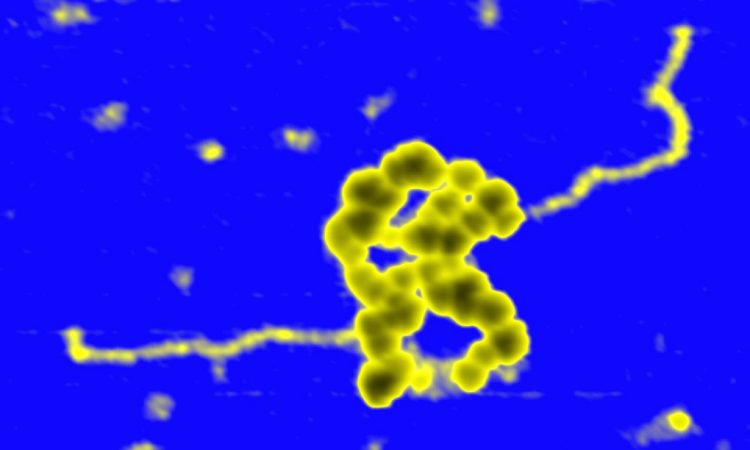
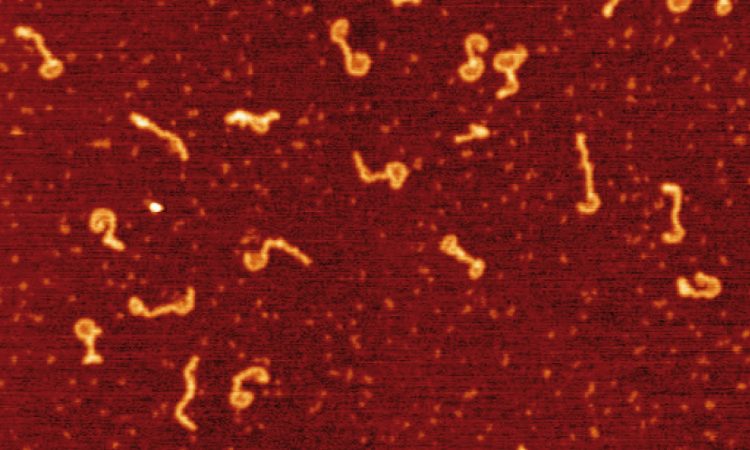
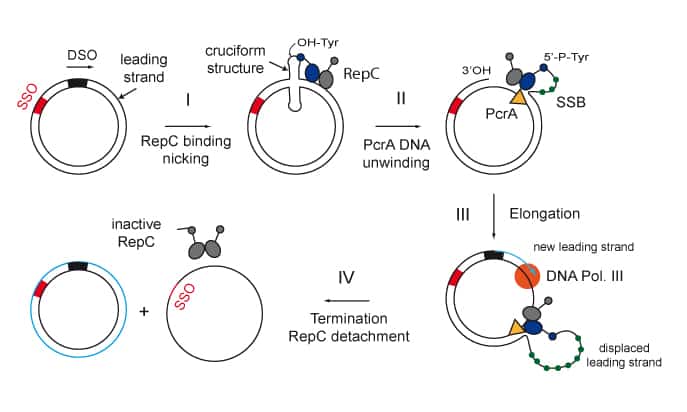
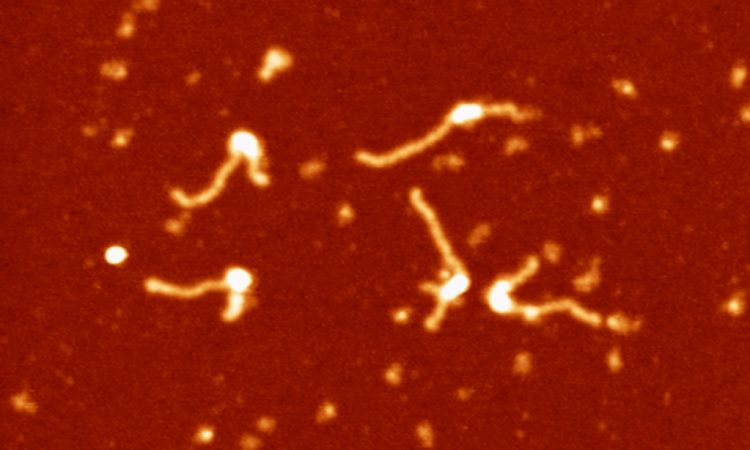
We are interested in studying double strand DNA break repair, Chromosome organisation and replication at the single molecule level using Atomic Force Microscopy, Magnetic and Optical Tweezers combined with Fluorescence, and standard biochemical techniques.
Welcome to the Moreno-Herrero Lab
The main interest of my group is to answer key questions in DNA-break repair, replication and Chromosome organisation using novel approaches based on single-molecule techniques. To do this, we develop our own instrumentation based on Atomic Force Microscopy imaging, single-molecule manipulation techniques such as Magnetic Tweezers combined with fluorescence, and establish strategic collaborations with research groups specialized on different biological systems. We are also interested in studying the mechanical properties of nucleic acids and their role in protein interaction. We investigate this from an experimental perspective using our single-molecule tools but also by all-atom molecular dynamics simulations.
We work at the National Center of Biotechnology (CNB), a research center part of the Spanish National Research Council (CSIC). The CNB is the largest CSIC institute with over 600 people working in a multidisciplinary environment that combines the latest technology in molecular biology, and structural and functional biology.
Check out our Openings section for opportunities to join the group to do the Master project, PhD or Postdoctoral research.
Research lines
NEWS & EVENTS
3rd Workshop on Advanced Nanobioscience
SBE “Manuel Rico” – Bruker Prize awarded to Fernando Moreno-Herrero
Fernando Moreno-Herrero has been awarded with the “Manuel Rico” – Bruker Prize of The Spanish Biophysical Society 2023 in recognition of his pioneering work in the development of molecular biophysics using Atomic Force Microscopy and Magnetic Tweezers in Spain. The jury highlights his pioneering biophysical studies on the molecular machinery involved in DNA repair, organization, and replication. This year's award has been jointly granted to Fernando Moreno-Herrero and Teresa Giráldez Fernández, professor at the University of La Laguna.